Here we describe the processes according to AquaBiota’s eDNA system.
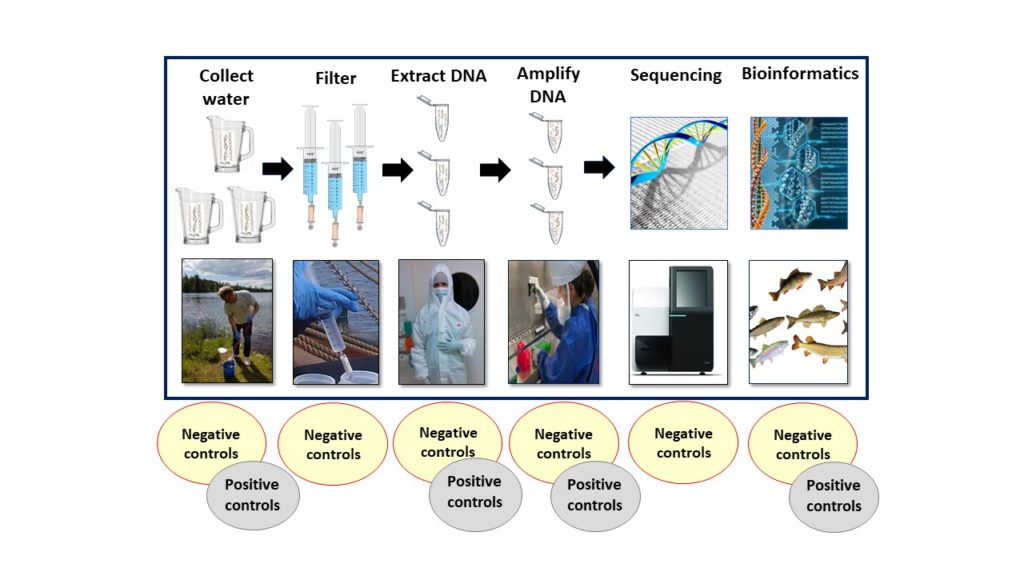
1. Collection of samples
Before the water samples are collected a sampling scheme is prepared in order to optimize the results, typically accounting for hydrology, sampling season and sample density. Prior to sampling all used equipment is sterilized in order to prevent contamination and cross contamination among samples. Water properties are measured during the sampling: temperature, pH and several other environmental parameters are registered, as they influence the amount of time eDNA remains in the water. This time can vary from a couple of days to a couple of weeks. Inadequate planning regarding where and how the samples should be taken can produce results that are unusable, which is why the planning for eDNA sampling is of great importance. The water is sampled in sterilised containers by trained personnel. It is crucial to use gloves and take precautions to avoid spreading of DNA. Water is filtered at the site through special filters, typically between 1-5 liter depending on the filter and the concentration of particles. The filters are of the same type used in purifying water from bacteria. At AquaBiota we save the filters and fix them in a fluid at the site. The filters are full of eDNA. The samples are then transported to the laboratories for analysis.
2. The extraction of eDNA
AquaBiota has, together with MoRe Research in Örnsköldsvik, established a sterile laboratory that is solely used for the extraction of eDNA. The work procedure is very similar to that of CSI, and DNA is extracted using the same principles used in a forensic laboratory. The researchers wear protective masks and full protective clothing during all work. This is of great importance as all risk of contamination must be avoided. Extracted DNA has a volume of 1/ 10 000 litres and this is sufficient for a number of analyses. DNA is saved in special designated spaces until further analysis.
3. Single and multiple species analyses
Depending on the question it is possible to detect one species at the time, or multiple species simultaneously, using molecular methods. Great Crested Newt is one example of an investigation we have performed as a single species. The investigations that for example are aimed at finding which fish species or amphibians are present in a given body of water we use the multiple species analyses, which means that you can analyse all species of fish and amphibians that are present in a sample at the same time.
3a. Single species analysis – barcoding
When you want to check whether a specific species, e.g. Great Crested Newt, is present in a pond, single species analyses are performed. The principle is based on that every living species excretes genetic material, or DNA. Certain parts of the DNA of an individual are unique to that individual, while other parts of the DNA are shared between all individuals of the specific species, and then there are parts of the DNA that is shared between all organisms within a taxonomic group. For single species analyses a short part of the DNA is selected that is unique for the specific species in question. Using large open access databases (https://www.ncbi.nlm.nih.gov/) the researchers can select a short sequence of the DNA that can be copied industrially. These short sequences are called primers. As the primer is species specific, it can only attach to DNA from the selected species. The result, i.e. a small part of DNA that the primer copies, is called a barcode, and can be compared to a barcode used on merchandise in a store. These primers, a drop of eDNA, an enzyme and salts are mixed in a test tube. The tube is placed in a machine that then produces numerous copies of the DNA for the selected species. This is known as PCR (Polymerase Chain Reaction) and is based on protein synthesis. The machines used are called qPCR and droplet PCR. While the sample is being run, a coloured curve will appear on a computer screen if the specific species is present in the sample. If the species is not present, the curve will not appear. The number of copies indicates the abundance present of the species in question. The results are available directly after the run.
3b. Multiple species analysis – metabarcoding using Next Generation Sequencing
Metabarcoding is an excellent method when you are looking to find out which species are present in a water sample using eDNA. With only one analysis, it is then possible to come up with thousands of species. The principle when using metabarcoding is the same as for barcoding for only one species. The primers for metabarcoding are produced so that they will select a part of DNA that is similar for all species within a group, for example all fishes, but the DNA that is sequenced varies between species and shows a unique code. As you are analysing multiple species simultaneously, this is called metabarcoding. DNA, primers, salts and enzymes are mixed, and the DNA is copied using PCR. To analyse the results the samples are run through a so called NGS (Next Generation Sequencing) analysis.
4. Multiple species analysis– bioinformatics
When the analysis is done, it results in enormous amounts of data, containing millions of different types of DNA. To organise the data and find out which species are present in a sample, different programming methods are used to sort the data. Enormous datafiles that correspond to several terabytes of data, are analysed using programs that separate the different DNA codes from each other. This process is known as bioinformatics. When the bioinformatics is done and the sequences sorted, a list is created of different DNA identities. These are then compared to different databases, and every unique piece of DNA receives an identity. The final result is an Excel file that specifies the species and how many copies of DNA is present for each species. It is then possible to say which species are present in a lake, and the relative abundance of each species. Metabarcoding from a water sample gives vast possibilities to analyse both previously known and unknown species that are present in a body of water in a very short time.
5. Multiple species analysis – ecological interpretation of the results
Specialists at AquaBiota go over the species lists and then write a report concerning the different species’ occurrence and their ecological significance. Unknown, rare and unwanted alien species can be found in the samples.
The post-data quality assurance includes a genetic review, where the similarity of species are compared within the analysed gene, to determine that the generated species list is correct. If this step is neglected, species can disappear due to false aggregation of species that have a high similarity in the analysed region. This can for instance happen among flatfishes when analysed within the commonly used 12S region. Beside this we also perform a geographic overview as well as cross primer comparison to ensure that the sequences get their correct species label.
As the field work with eDNA sampling is considerably faster than conventional methods, large areas can now be sampled in a short period of time. Investigations using eDNA that are repeated at regular time intervals are very useful to verify environmental changes over time, or if species disappear due to different actions such as dam constructions or factory constructions near streams. AquaBiota has during the summer of 2017 carried out several metabarcoding projects concerning fish, and the results have corresponded well with aggregated data from several years of exploratory fishing. eDNA gives a considerably better view of species’ occurrences than conventional inventory methods, such as exploratory fishing, electrical fishing or interviews with the local population.